align_masks.html
Aligning the masks
The first script we need to run in order to make a spectral map is align-mask, which uses the comparison lines to calculate the large-scale offsets and adjust the dewoff file accordingly. To edit the parameters for align-mask, we use the editpar routine:
editpar align-mask type q to end SEARCHBOX 10 half width of search range MAGFACTOR 100 magnification factor for offset vectors LAMFILE somelines.dat line list file NAVER 1 number of neighboring points to average THRESHOLD 0.100000 minimum flux ratio spot to background SIGLIMIT 10.000000 line rejection threshold MAXFLUX 65000.000000 maximum flux WINDOWSIZE 7.000000 width of PGPLOT window in inches Change parameter:For these data, I put the lines at 4921.93, 5015.68, and 5400.56 Angstroms in somelines.dat, because those lines are relatively isolated and not saturated. Avoid using more than 3 lines, as this may cause confusion in align-mask. Then, we run the command:
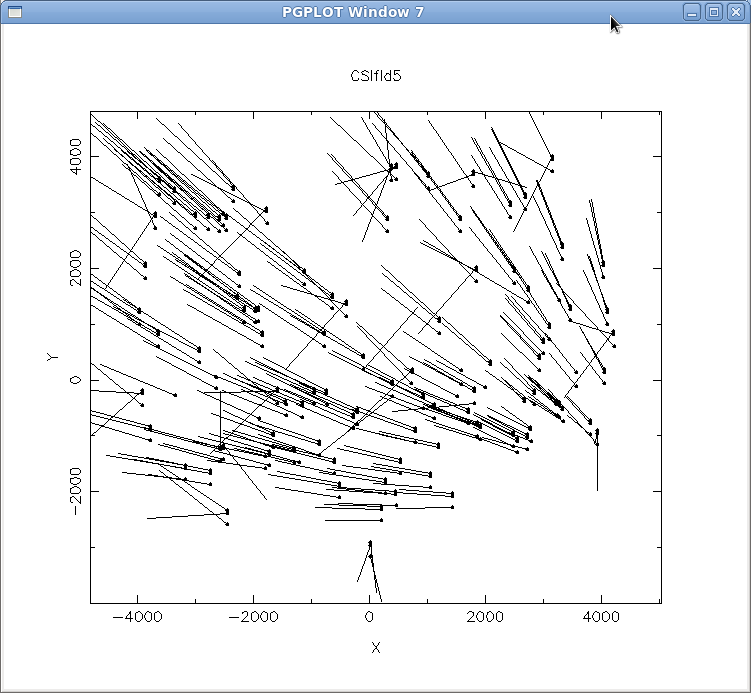
align-mask -o CSIfld5 -f ift0012 parameter file ./align-mask.par found Processing "CSIfld5.SMF". ** Unknown keyword "DREF" ignored. ** Unknown keyword "DATE" ignored. ** Unknown keyword "XFWH9HRN1EPQX4OR4HP4KGD_QYMVUM FXKNVSS7QHPASN9ETBCADLCWZA_W5NU23IMLDY6Z" ignored. Using E2V1 distortion corrections Delta=(-10.01 5.48) Sigma=(3.35 5.08) Clipping out (1532.45,-343.53) Clipping out (1534.07,-546.96) Delta=(-10.02 5.48) Sigma=(3.33 5.06) Clipping out (-2270.69,1252.93) Clipping out (425.03,2647.53) Clipping out (425.70,2435.20) Delta=(-10.03 5.48) Sigma=(3.31 5.06) Clipping out (-2518.94,3031.25) Clipping out (-1780.39,-1325.13) Clipping out (-3346.16,-229.54) Clipping out (-3346.76,-485.34) Clipping out (2674.58,-582.58) Clipping out (1204.60,-1409.91) 297 matches found. sigma= 12.74 pixels Continue? y Average scale = 0.99869 Reset scale? y Average rotation angle = -0.013 Reset angle? y Average X,Y Offsets = -9.9 6.2 pixels. Apply shifts? y Creating new dewar offset file ift0012.dewoffA PGPLOT window will pop up with a vector plot of the calculated offsets. As you can see in Figure 3, align-mask detects the large-scale shifts and applies these corrections to the dewoff file. If you do not see a large-scale shift such as this for other data, then most likely you need to either increase your SEARCHBOX or adjust the dewoff file manually using the adjust-offset routine. Since we see a noticeable bulk shift, however, we can proceed to adjust the scale and angle, and apply the shifts to the dewoff file.
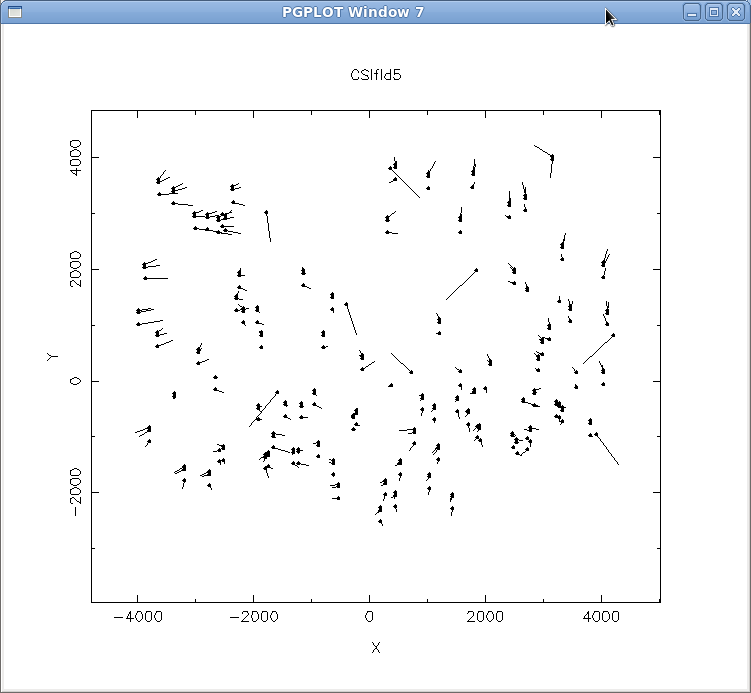
The output of align-mask in this case is ift0012.dewoff. Edit CSIfld5.obsdef to use ift0012.dewoff and then rerun align-mask. After a few iterations, you'll notice that the offset "sigma" has gone down to less than 2 pixels (see Figure 4), which means we now have a good enough dewoff to move on.
Next: Constructing the Spectral Map Up: Running Sample IMACS data Previous: Spot-check the features Contents
Edward Villanueva 2014-08-27