spot-check.html
Spot-check the features
Before doing any wavelength calibration, it is beneficial to check how good your initial dewar offset file is, since the initial offset may be too large for align-mask to detect. To do this, we can run the spectral-lines program with the full HeNeAr.dat line list to produce a new file with the X and Y coordinates of the comparison lines for each slit:
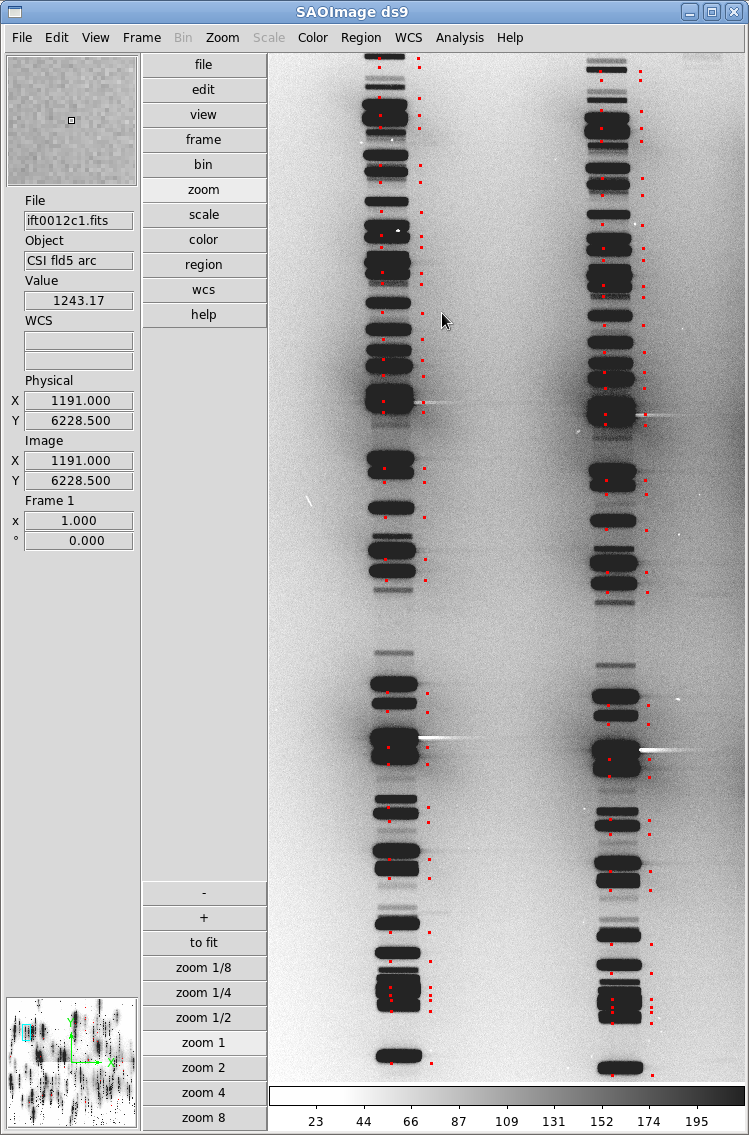
spectral-lines -o CSIfld5 -l HeNeAr.dat -b 1 -eThen we use display8 and tvmark in IRAF to display a mosaic of the lamp frames and overplot the positions from the resulting CSIfld5.xy file:
ecl> display8 ift0012 1 ecl> tvmark 1 CSIfld5.xy color=204As you can see in Figure 2, the initial positions aren't too far off, so we can proceed to the next step of aligning the masks. If they don't seem close at all, you will need to use the adjust-offset routine to adjust the dewoff file. You may also need to alter the Disperser Misalignment with defineobs if you see a rotation offset in the positions.
|
Next: Aligning the masks Up: Running Sample IMACS data Previous: Setting up observation definition Contents
Edward Villanueva 2014-08-27
